Dispersion (R,Python, Excel, JASP)
Red means that the page does not exist yet
Gray means that the page doesn’t yet have separation of different levels of understanding
Orange means that the page is started
In this website you can choose to expand or shrink the page to match the level of understanding you want.
- If you do not expand any (green) subsections then you will only see the most superficial level of description about the statistics. If you expand the green subsections you will get details that are required to complete the tests, but perhaps not all the explanations for why the statistics work.
- If you expand the blue subsections you will also see some explanations that will give you a more complete understanding. If you are completing MSc-level statistics you would be expected to understand all the blue subsections.
- Red subsections will go deeper than what is expected at MSc level, such as testing higher level concepts.
To understand distributions such as the normal distribution, it’s helpful to clarify some more basic concepts around how data is dispersed or spread.
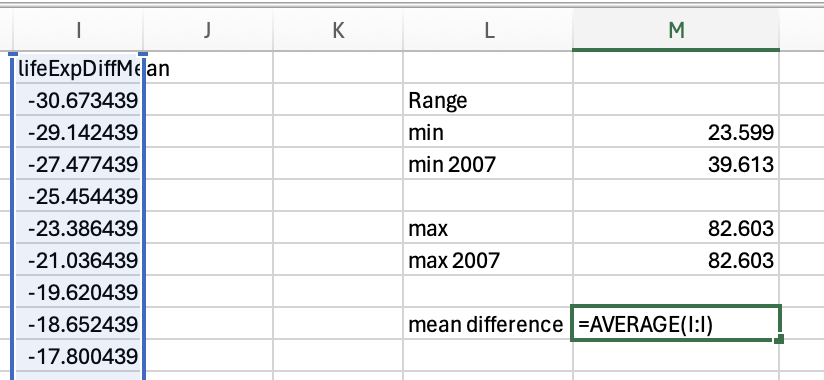
Range
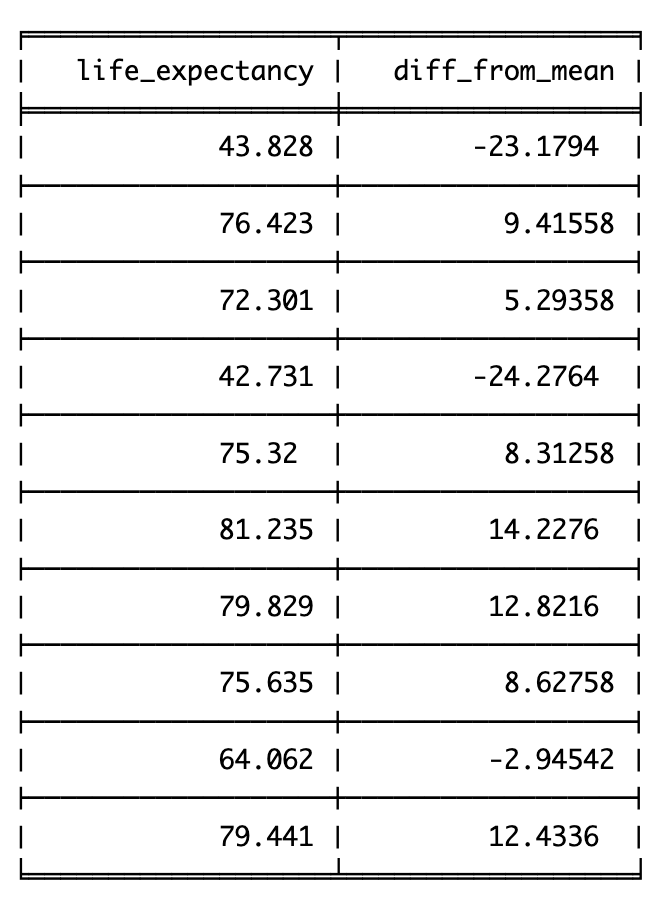
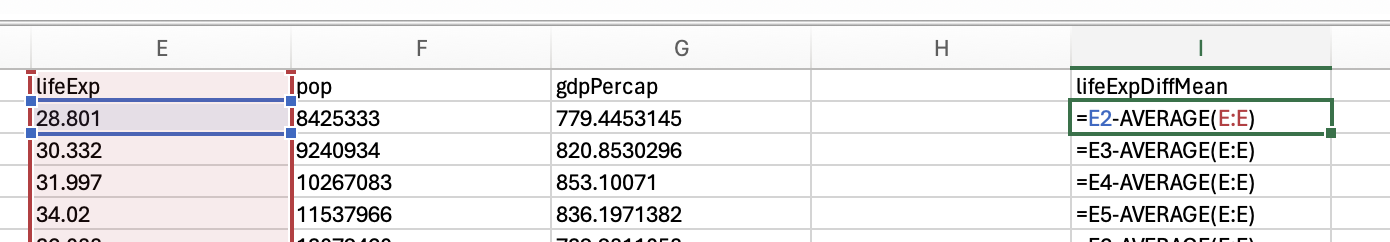
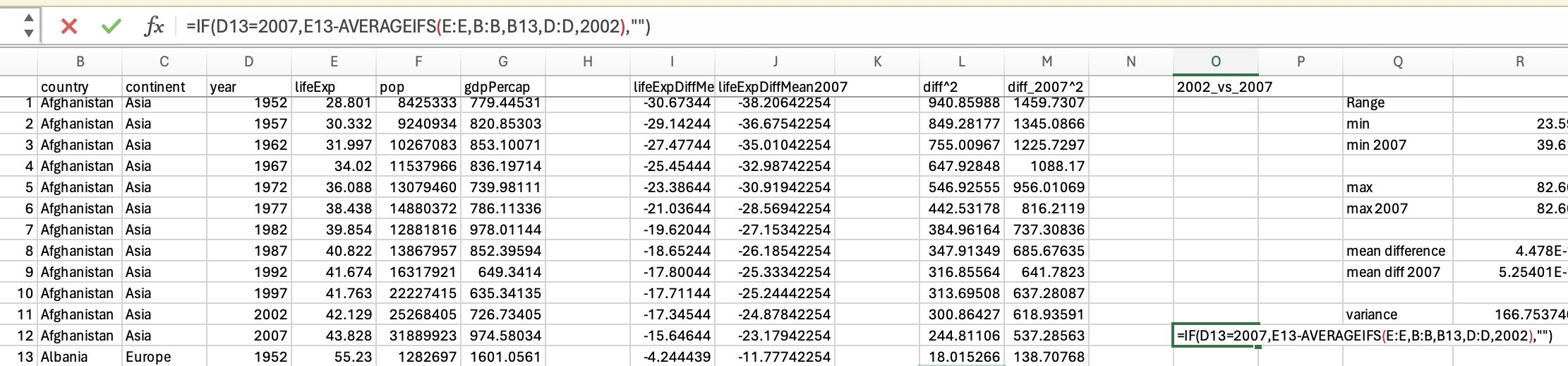
Range simply captures the min(imum) and the max(imum) values. Lets look at the min and max for the life expectancy data from 2007:
# load the gapminder data
library(gapminder)
# create a new data frame that only focuses on data from 2007
gapminder_2007 <- subset(
gapminder, # the data set
year == 2007
)
min(gapminder_2007$lifeExp)[1] 39.613max(gapminder_2007$lifeExp)[1] 82.603# load the gapminder module and import the gapminder dataset
from gapminder import gapminder
# create a new data frame that only focuses on data from 2007
gapminder_2007 = gapminder.loc[gapminder['year'] == 2007]
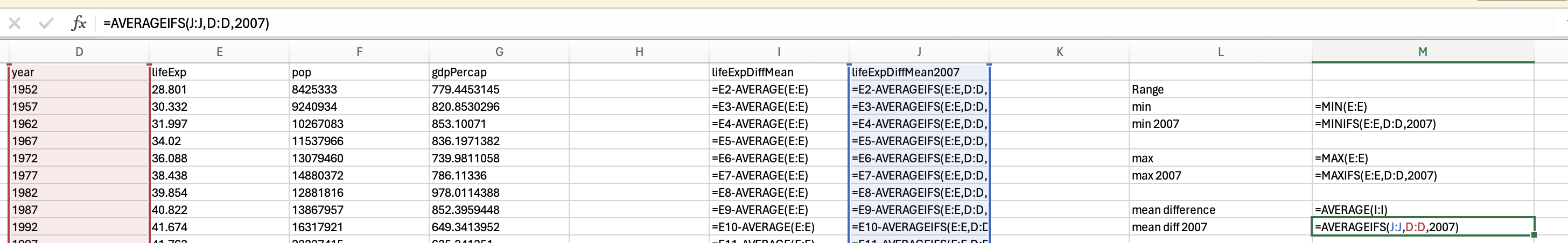
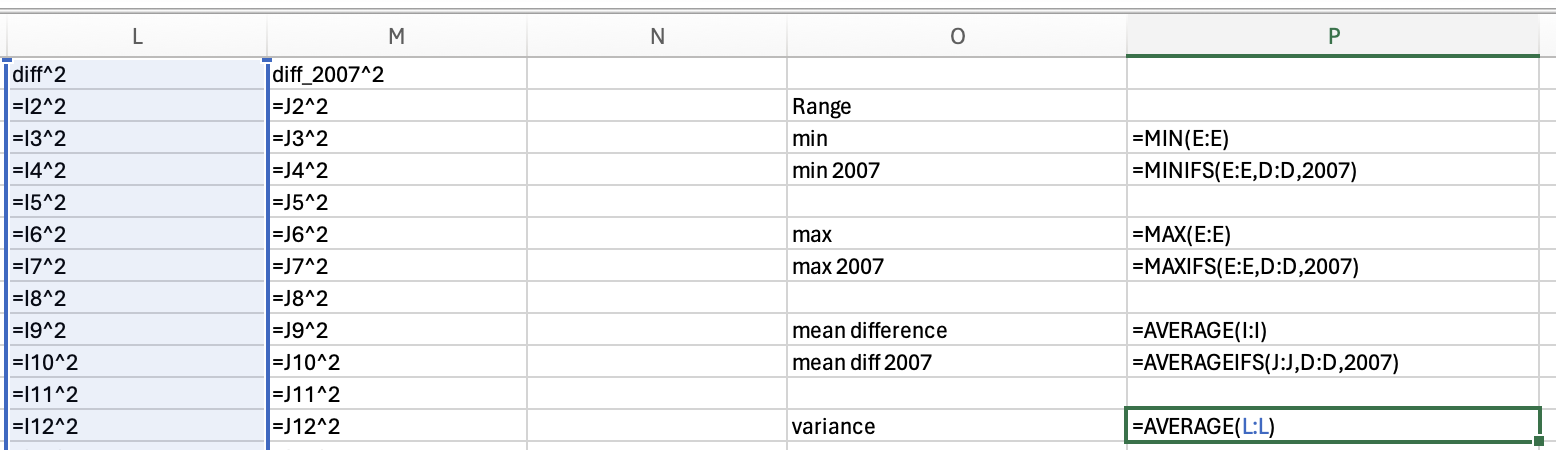
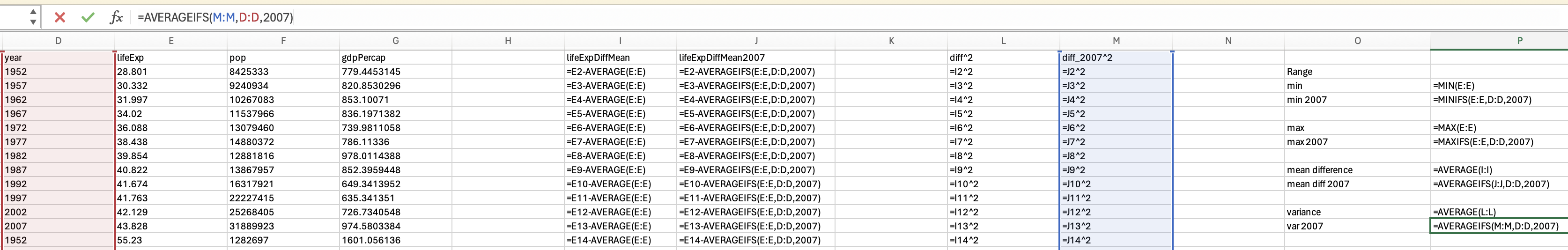
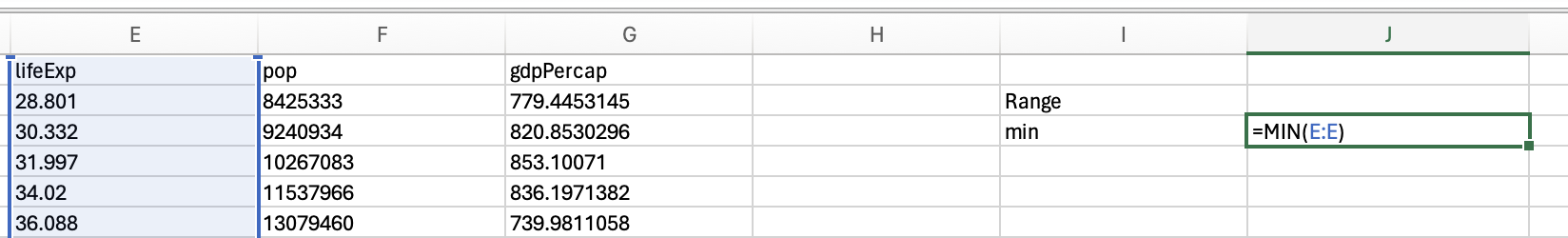
gapminder_2007['lifeExp'].min()39.613gapminder_2007['lifeExp'].max()82.603You should be able to access an excel spreadsheet of the gapminder data here. To get the minimum you can use the min formula:
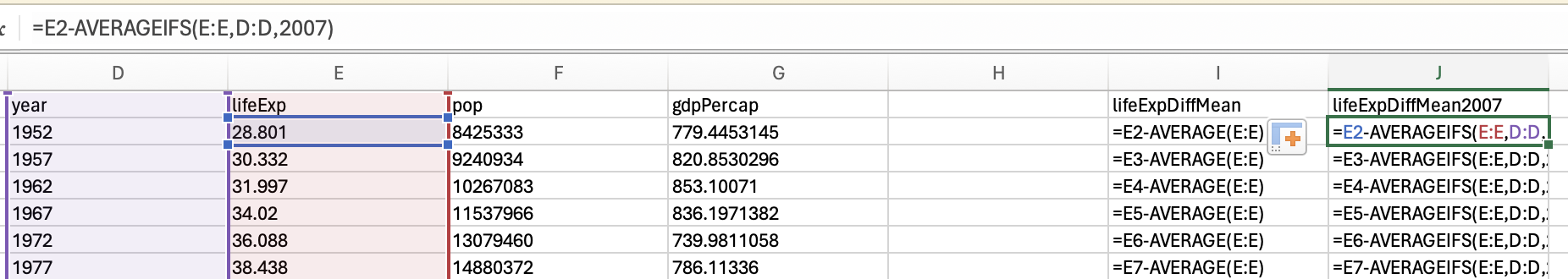
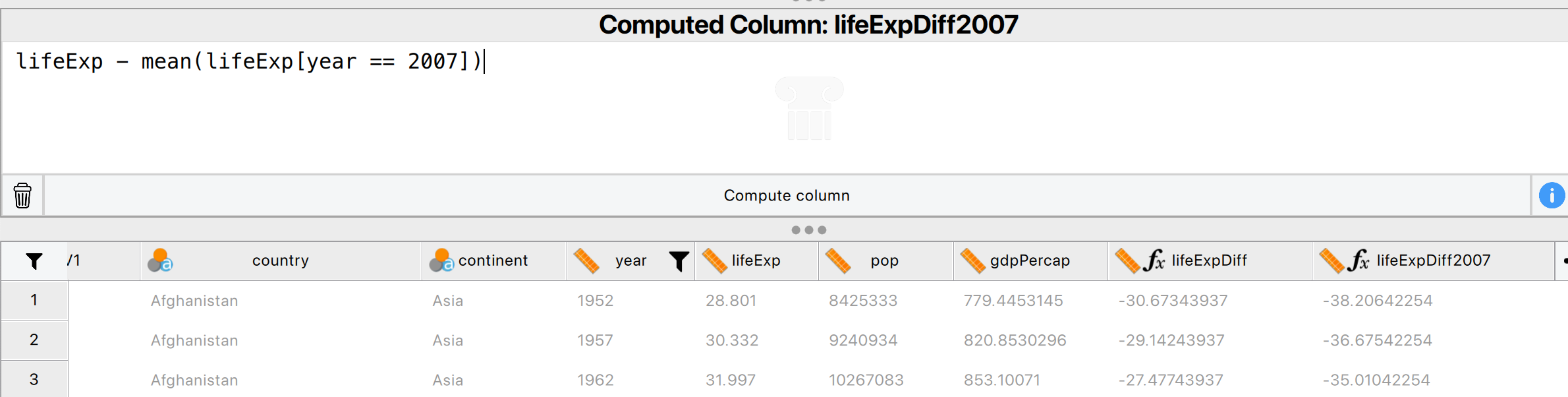
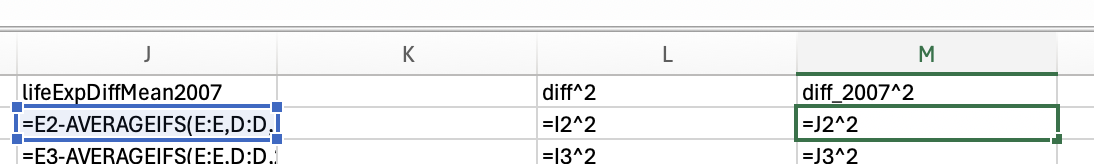
However, we want the minimum life expectancy in 2007. To do this, we need to use the minifs formula (which uses similar logic to averageifs):
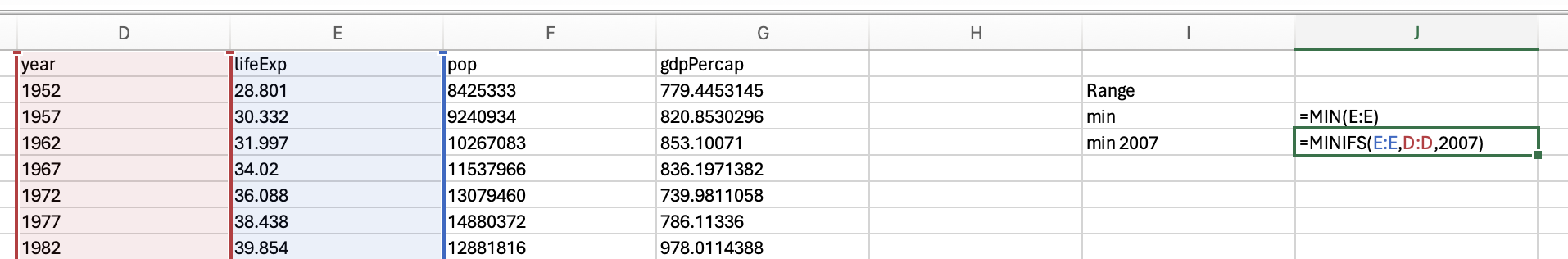
Which would give us 39.613.
Just use the max function to get the maximum life expectancy across all years:
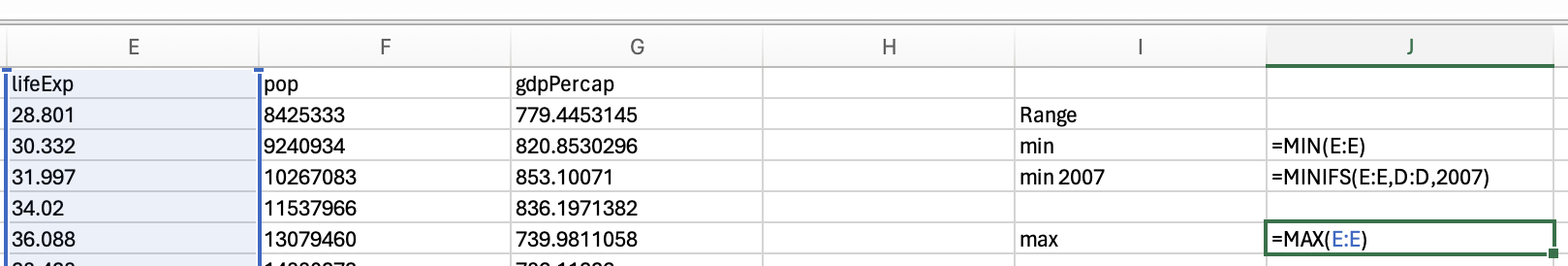
and the maxifs function to get the max life expectancy in 2007:
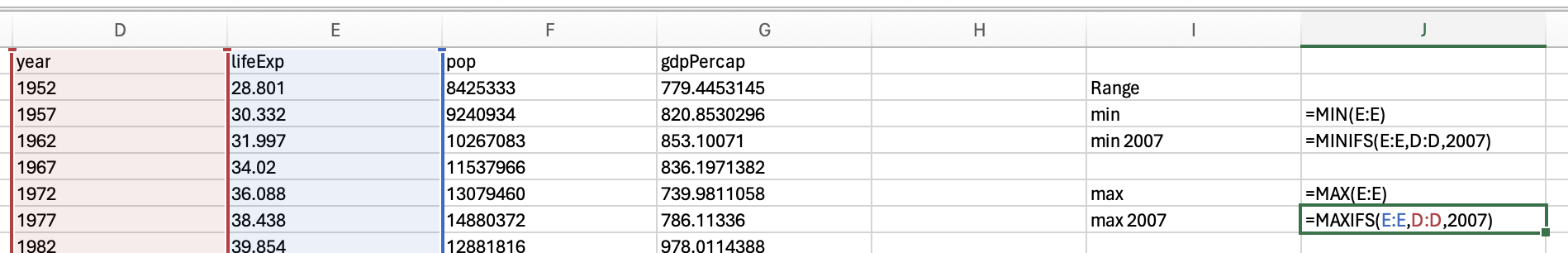
82.603
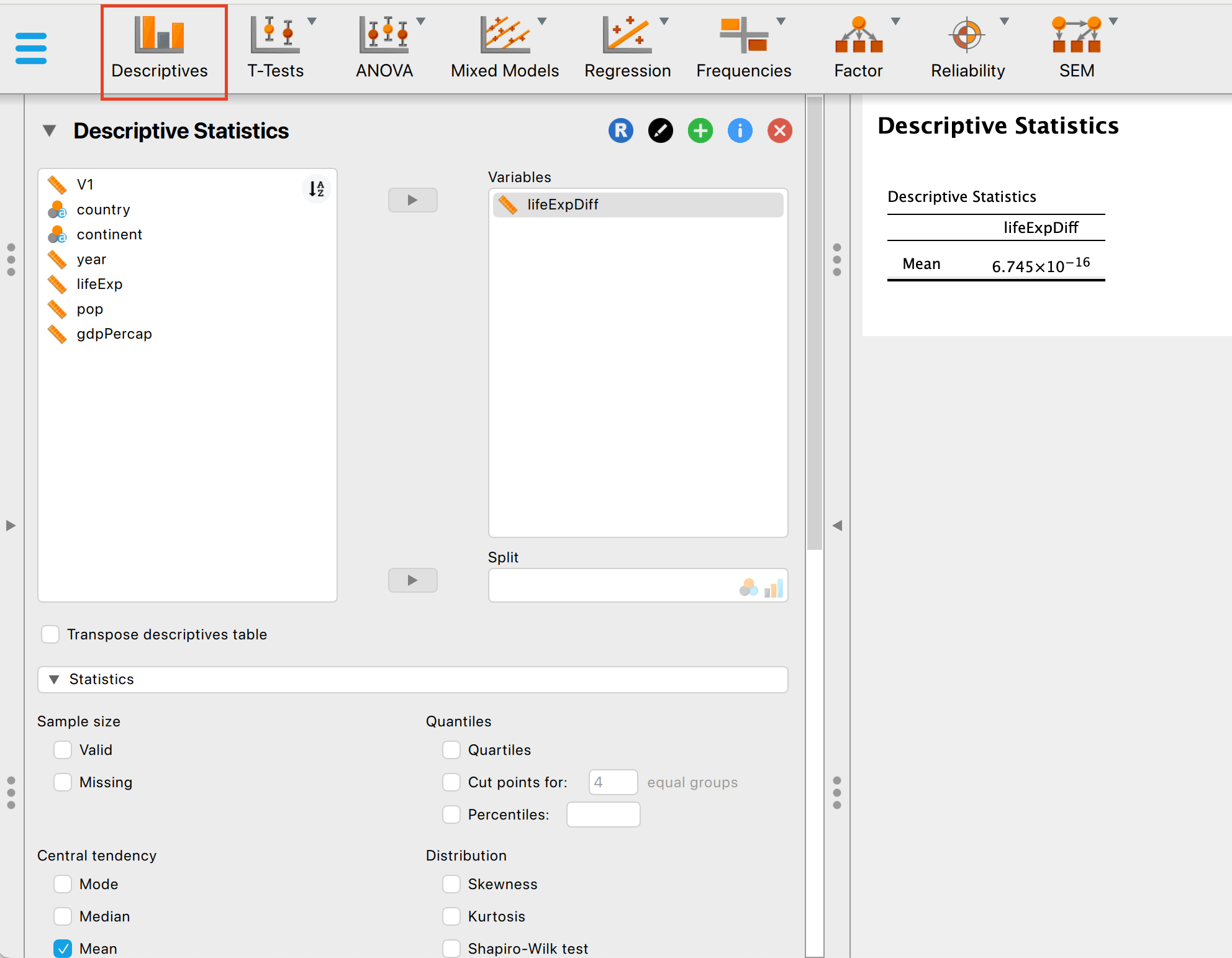
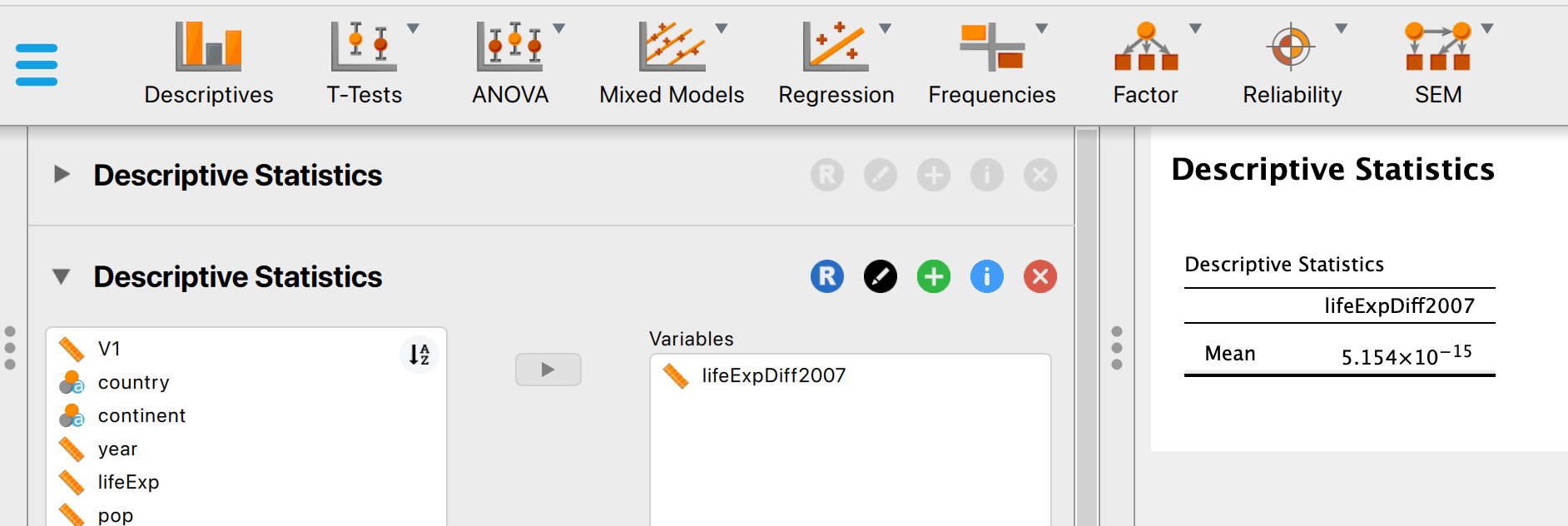
To get the max and minimum values across the whole data, you can use the descriptives interface:
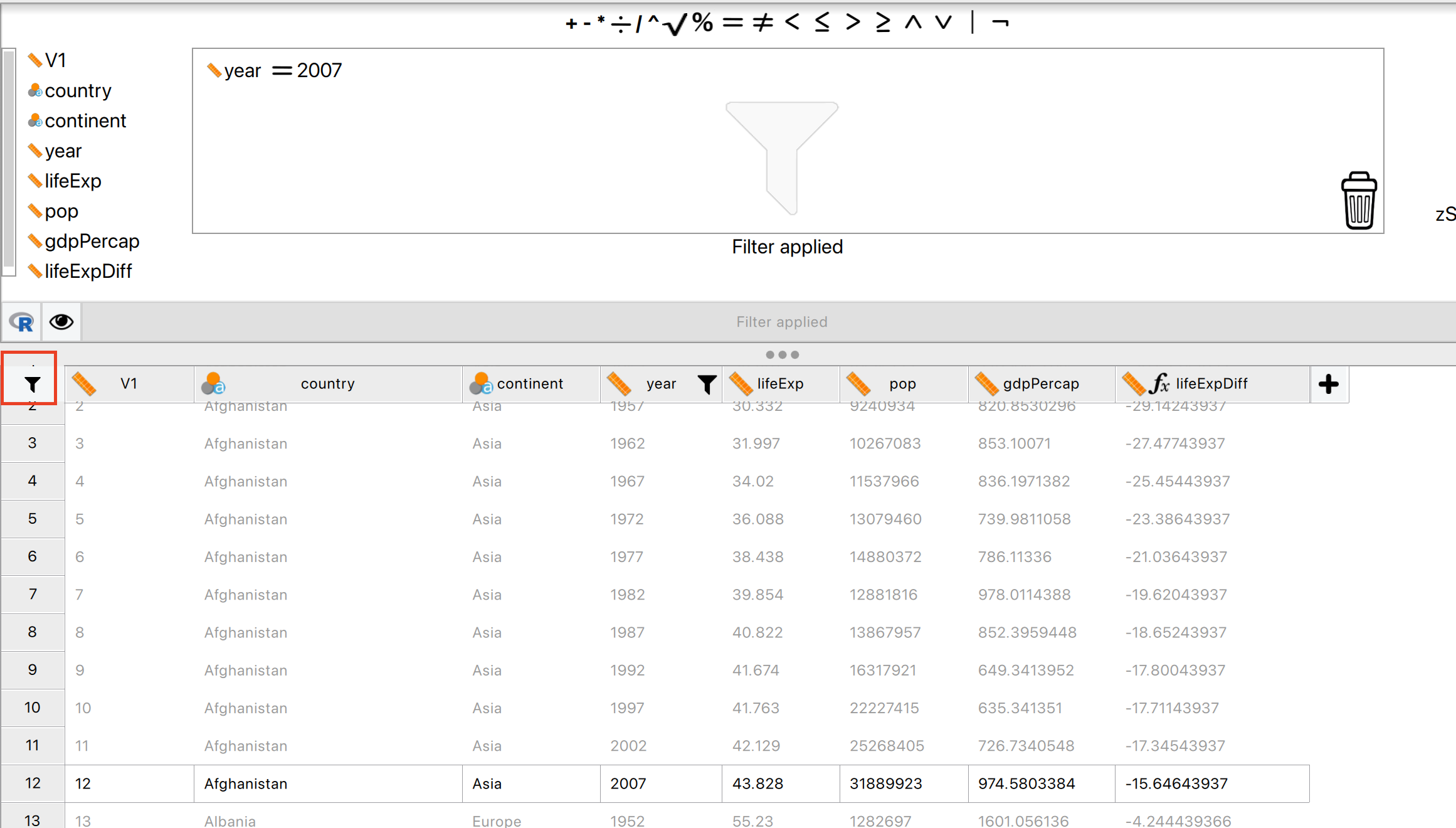
But if you want the minimum and maximum values for just 2007 data, you will need to apply a filter first:
So the range for life expectancy in 2007 was between 39.613 and 82.603.
Variance
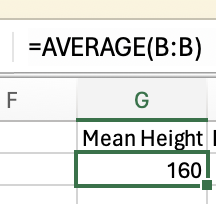
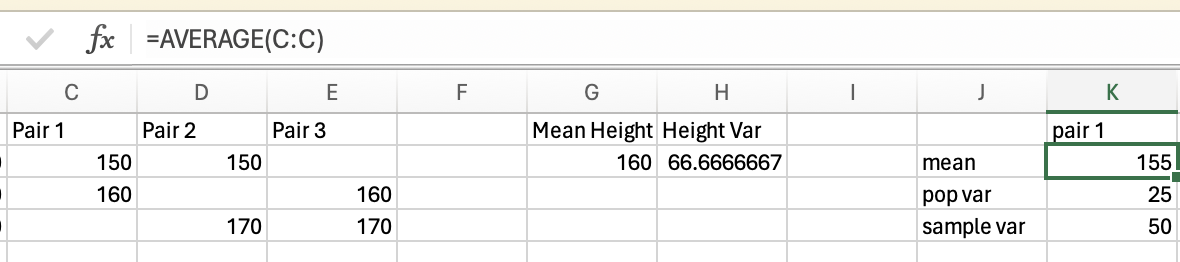
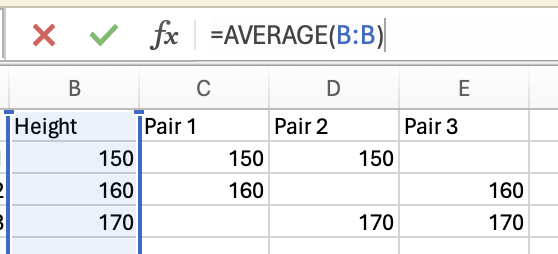
Variance is how much the data varies or fluctuates around a mean. If there’s a lot of variance around a mean then that suggests the mean isn’t very representative of your data. Let’s imagine you have 2 sets of 5 pets, and you want to know what the mean height is, and how much variance there is around the mean.
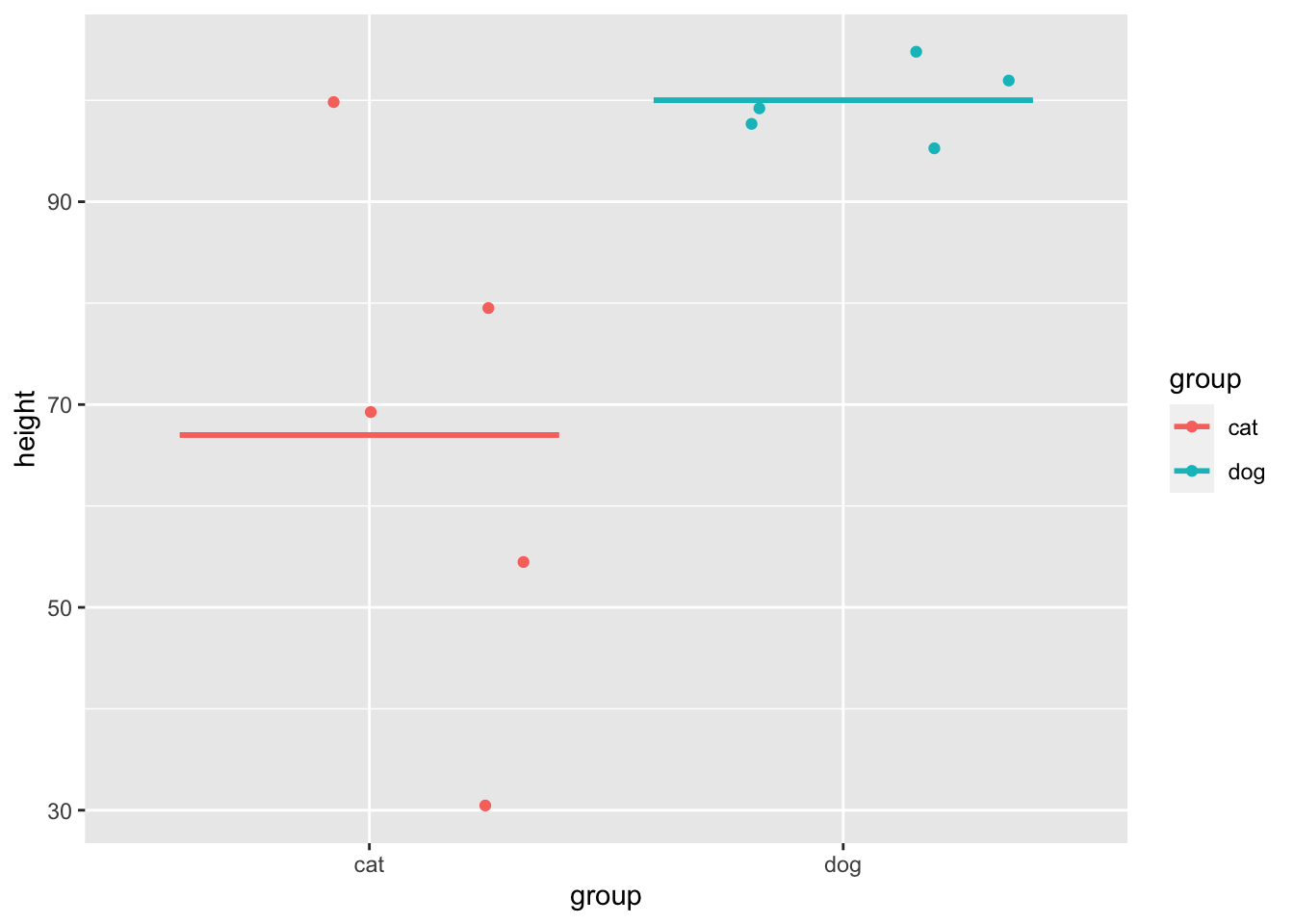
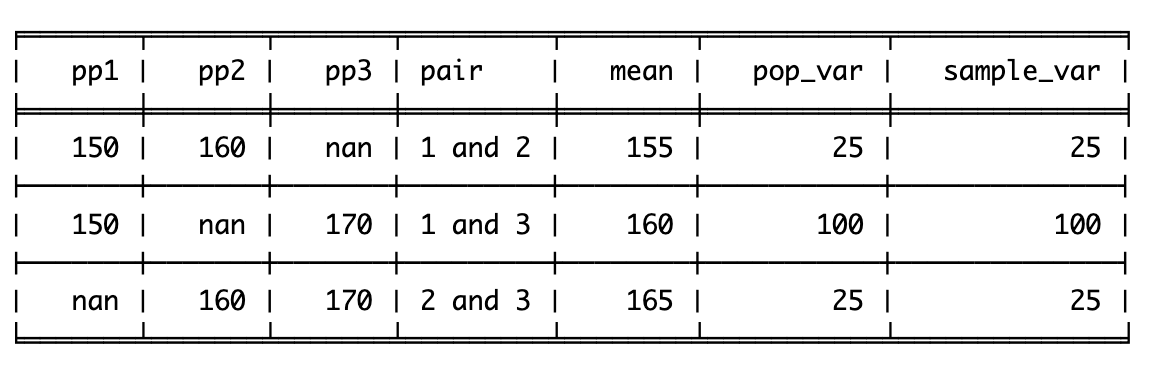
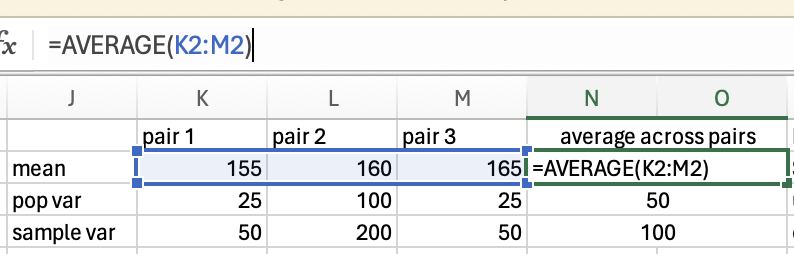
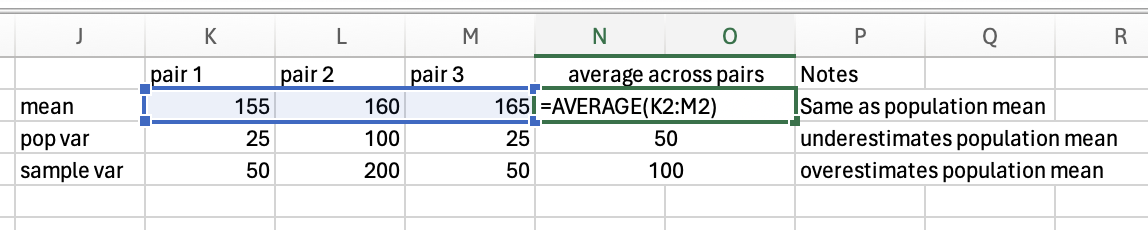
The variance in height among cats is higher than among dogs, as there is very little fluctuation (around the mean) in the dog data, but a lot of fluctuation in height within the cats. Population variance refers to variance when you have measured all items/animals/people within a population. For example, if there were only 5 cats and 5 dogs in the world, then you could calculate the population variance using the height of all 10 of the animals. However, there are quite a lot more than 10 cats and dogs in the world, so it would not be realistic to measure all of their heights. Thus you would calculate the sample variance instead on a sample of cats and dogs (e.g. 5 of each). Importantly, the calculation for the population vs. sample variance are slightly different (see degrees of freedom for an explanation of why), and you will almost always use the calculation for sample variance (as you rarely have every member of a group that you are investigating).
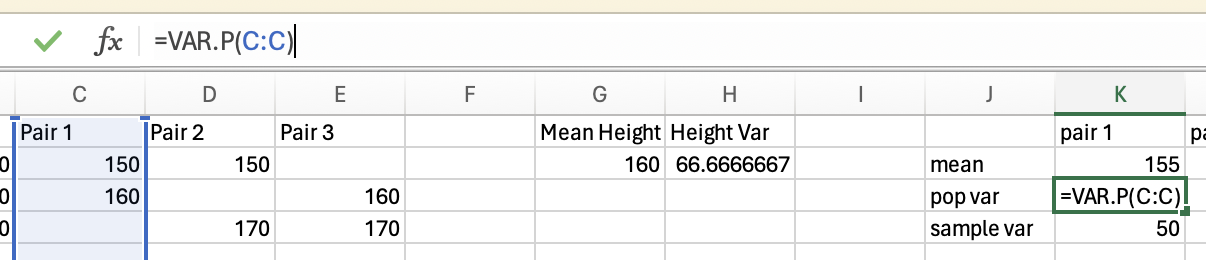
Population Variance
The population variance is a summary statistic of how much data varies around the mean. It does this by calculating the mean squared difference between each data point and the group mean.
Using our pet example from above, we would calculate the average squared distance for cats and dogs by first capturing the difference between the animal’s height and their group mean:
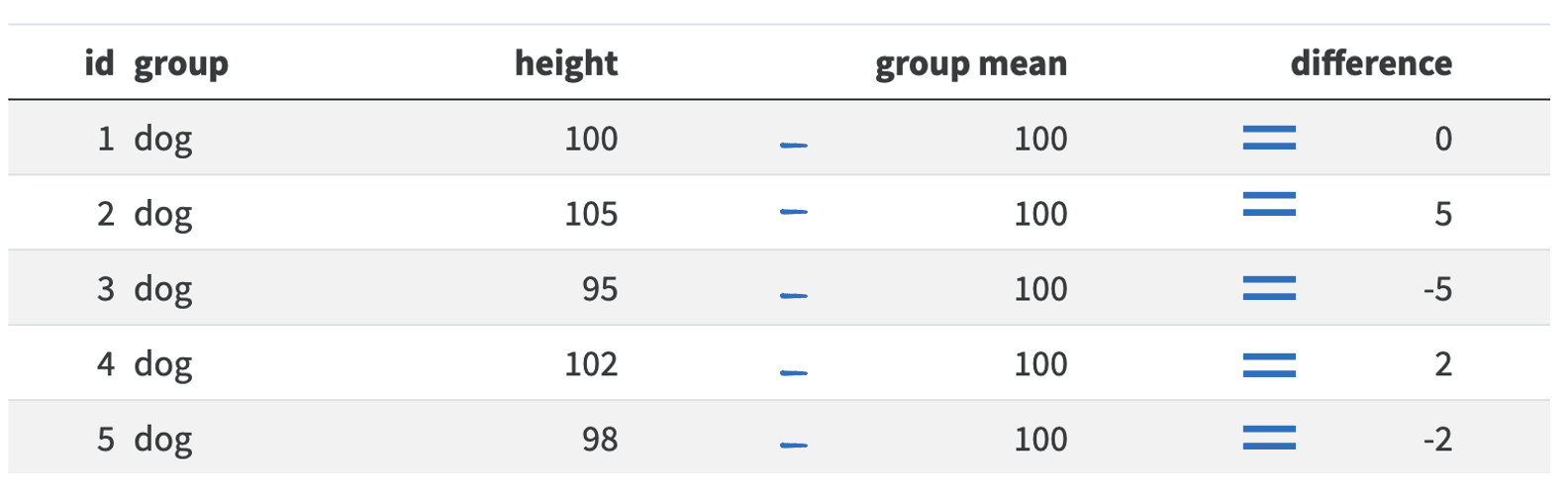 So for dog 1 there was no difference between its height and the mean height for dogs. Dog 2 is 5 centimeters taller than the mean, so the difference is 5. However, dog 3 is 5cm shorter than the mean, so its difference is -5cm. If we calculated the mean before squaring these differences, we would get 0 (\((0 + 5 - 5 + 2 - 2)/5 = 0/5 = 0\)). So we need to square all the numbers to make them positive:
So for dog 1 there was no difference between its height and the mean height for dogs. Dog 2 is 5 centimeters taller than the mean, so the difference is 5. However, dog 3 is 5cm shorter than the mean, so its difference is -5cm. If we calculated the mean before squaring these differences, we would get 0 (\((0 + 5 - 5 + 2 - 2)/5 = 0/5 = 0\)). So we need to square all the numbers to make them positive:
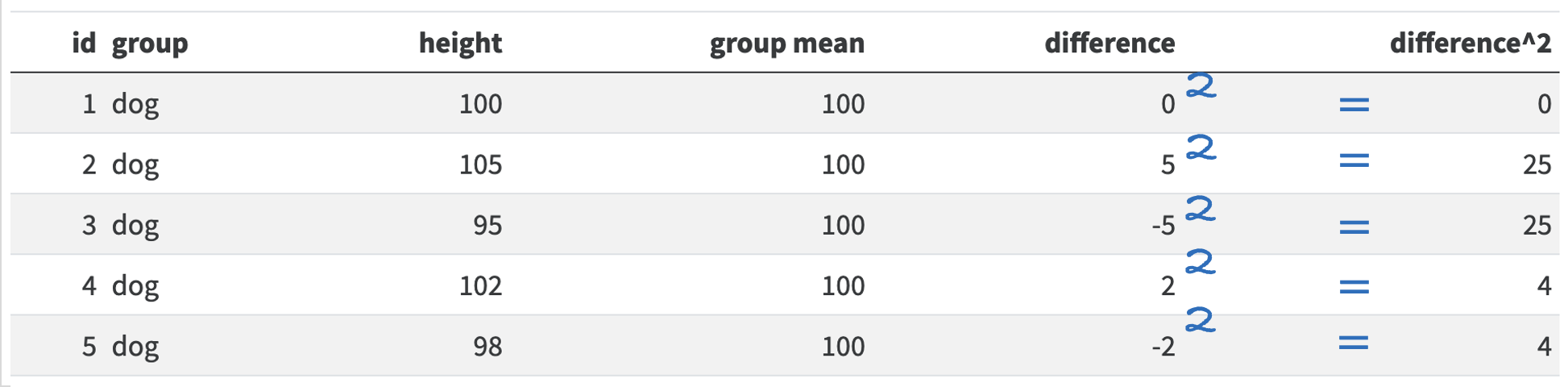 And then we can average of these squared numbers to get our variance score:
And then we can average of these squared numbers to get our variance score:
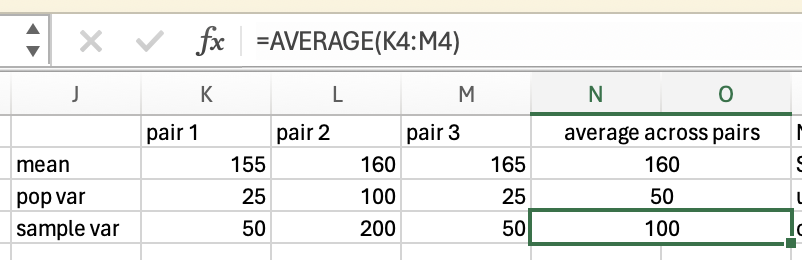
\(variance = (0 + 25 + 25+ 4+4)/5 = 58/5 = 11.6\)
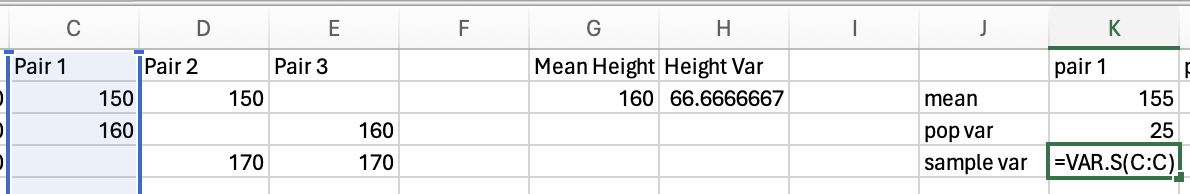
Sample variance
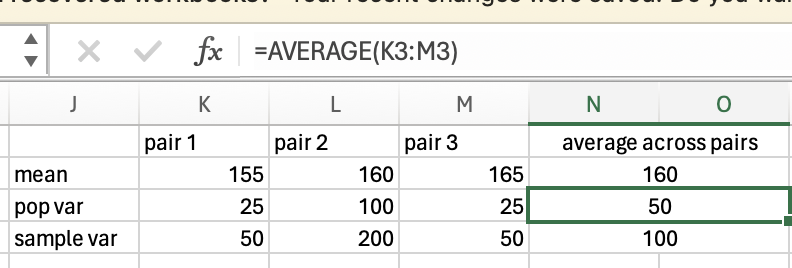
To calculate the variance for a sample of participants, rather than the entire population of the group you’re measuring, you need to correct for the fact you are only measuring a sample, not the entire population (this is explained further in the degrees of freedom section below). This means that instead of calculating the mean of the differences squared (see above), you need to do a slightly different calculation. The normal calculation for mean is the sum of all scores divided by the number of items you have. For sample variance, you correct for uncertainty by dividing by N-1 instead of dividing by N.
Let’s apply this to the dog example above:

Instead of dividing the differences squared (58) and by the number of items (5), we divide 58 by the number of items minus 1 (4):
\(58/(5-1) = 58/4 = 14.5\)
Degrees of Freedom
Degrees of freedom are used to address how analyses of samples can sometimes underestimate dispersion (e.g. variance) that would be found in the population. Degrees of freedom are applied by adapting certain formulas that would normally include an N value (number of items/people/data points) to increase the estimate, e.g. estimated variance. For example, let’s compare the formulas for population vs. sample variance:
| Population variance | Sample Variance |
|---|---|
| \[ \sigma^2 = \frac{\sum((x_i- \bar{x}{} )^2)} {N} \] | \[ S^2 = \frac{\sum((x_i- \bar{x}{} )^2)} {N - 1} \] |
In the population formula you divide by \(N\), in the sample formula you divide by the degrees of freedom \(N-1\). Note that the sample variance formula has \(N-1\) as the denominator (i.e. bottom half), which will increase the estimate compared to having just \(N\) as the denominator.
Standard deviation (SD)
Standard deviation is the square root of the variance. This takes into account that that the variance includes the square of the difference between the individual values and the mean:
| Population | Sample | |
|---|---|---|
| Variance | \[ \sigma^2 = \frac{\sum((x_i- \bar{x}{})\color{Black}{^2}\color{Black})} {N} \] | \[ S^2 = \frac{\sum((x_i- \bar{x}{} )\color{Black}{^2}\color{Black})} {N - 1} \] |
| SD | \[ \sigma = \sqrt\frac{\sum((x_i- \bar{x}{})\color{Black}{^2}\color{Black})} {N} \] | \[ S = \sqrt\frac{\sum((x_i- \bar{x}{} )\color{Black}{^2}\color{Black})} {N - 1} \] |
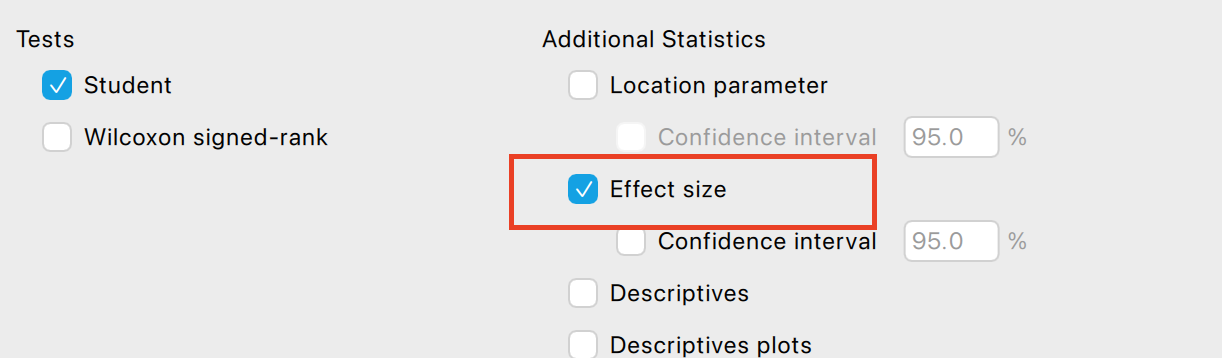
Effect size
Effect size is basically what it sounds like, it is a measure of how big the effect you are investigating is. You may find a difference between participants or conditions, but effect size calculations give you a sense of whether these are big or small effects. We will use Cohen’s D as an example of an effect size calculation to illustrate this issue.
Cohen’s \(d\) is used to capture the effect size in three situations:
Comparing a set of data against a single value (e.g. how much is life expectancy higher than 55 years)
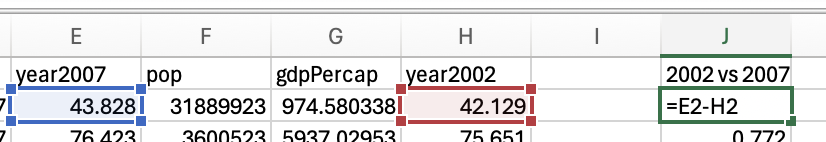
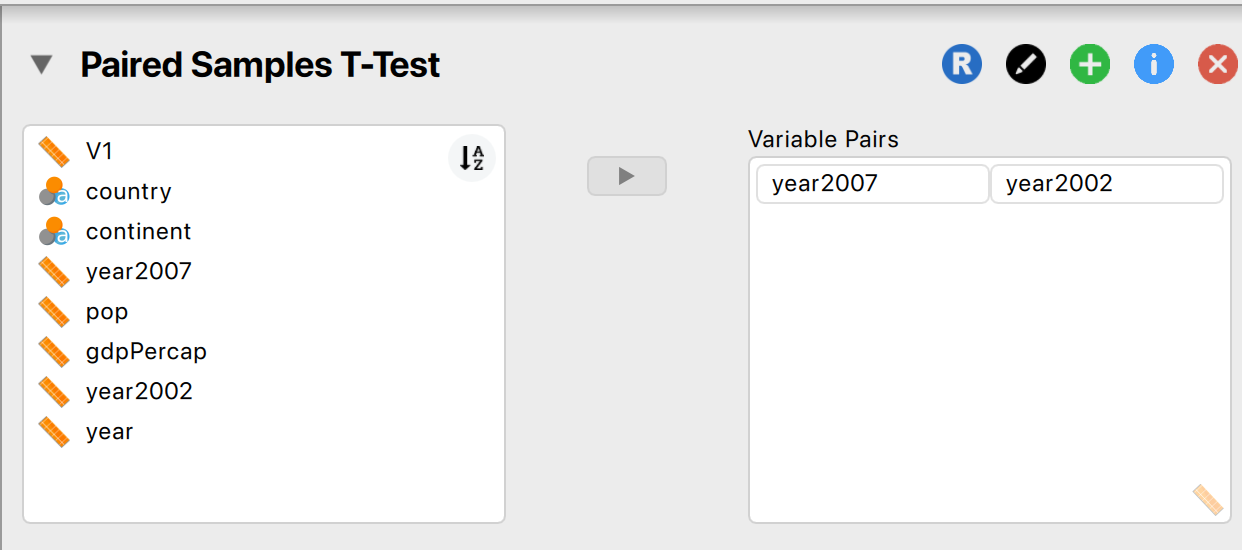
Comparing two conditions (within-subject; e.g. how much has life expectancy gone up or down between 2002 and 2007)
Comparing two groups of participants (between-subject; e.g. how different is life expectancy between 2 countries)
The general benchmarks for how big or small a Cohen’s \(d\) value are as follows:
.01 is very small (Sawilowsky 2009)
.2 and below is small (Cohen 2013)
.5 is medium (Cohen 2013)
.8 is large (Cohen 2013)
1.2 is very large (Sawilowsky 2009)
2 is huge (Sawilowsky 2009)
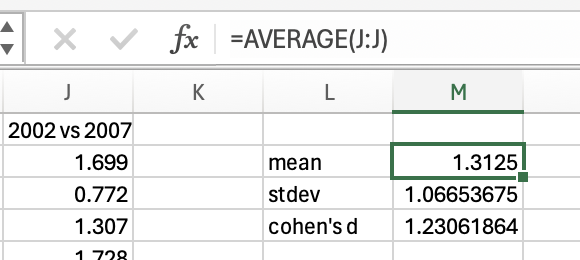
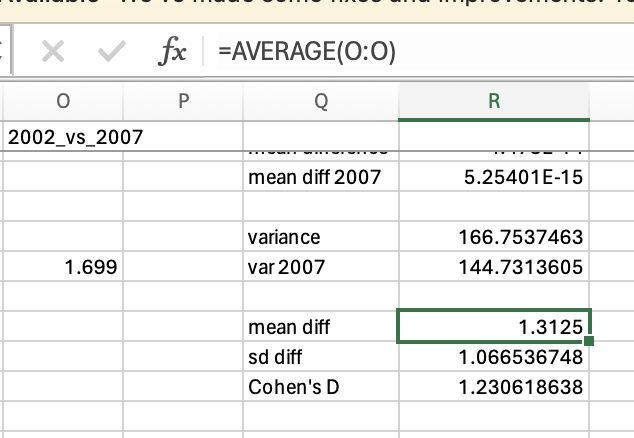
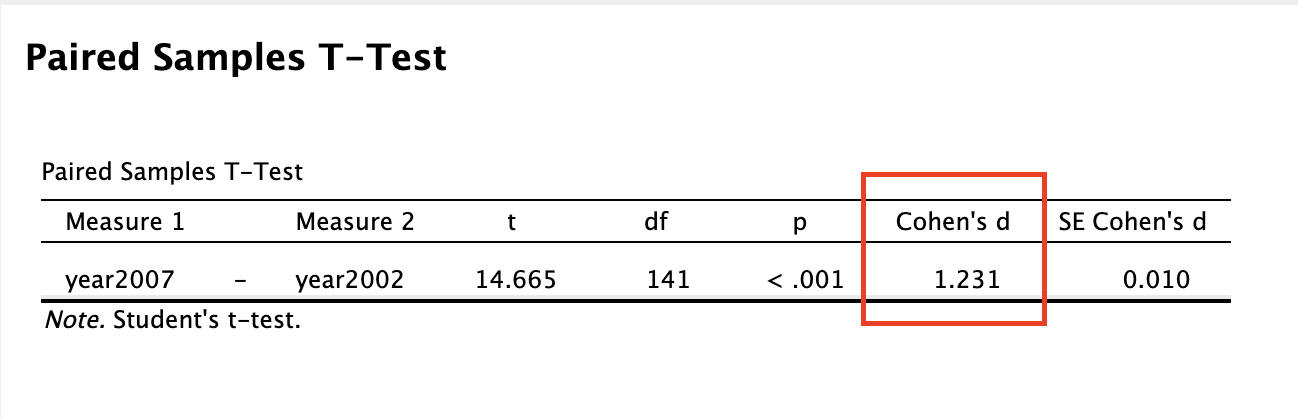
Now, you may be thinking that we already had insight into the effect size simply by comparing means, in which we can see that life expectancy has gone up by 1.31 years between 2002 and 2007. That’s true, and depending on your research question you may want to acknowledge whether the change is meaningful (i.e. is one year’s higher life expectancy a big deal?). However, you might not say an average increase of 1.31 years in life expectancy is a large effect if the change of life expectancy is very inconsistent between countries (e.g. some countries life expectancy decreased). Effect size calculation takes into consideration the how consistent the data is and so gives you further insight than a simple mean difference. The large effect size (1.23) confirms that there is a large effect of time on life expectancy, that there is a big difference relative to the general variation in the data.
Note that many tests described later have their own effect size calculations associated with them:
| Test | Effect Size Unit/Test |
|---|---|
| T-tests | Cohen’s \(d\) |
| Correlations | \(r\) |
| Regression | \(R^2\) |
| ANOVAs | \(\eta\) |
Question 1
True or False: Using degrees of freedom (N-1) rather than N controls for bias
Question 2
Which of the following can be negative?